Prof Joep Lange was a pioneer of antiretroviral treatment (ART) in Africa. With PharmAccess he rolled out ART programs in Africa’s private sector already in 2001. To date, access to ART in sub-Saharan Africa has reached unprecedented levels:
“The best should never be the enemy of the good. For scaling diagnostics of diseases, it is better to implement relatively ‘sloppy’ assays with large population coverage than offer a 99,99% gold standard assay for privileged individuals.”
Tobias Rinke de Wit

However, ART programs are at risk again today. As stated by UNAIDS:
‘The COVID-19 pandemic has seriously impacted the AIDS response and could disrupt it more. A six-month complete disruption in HIV treatment could cause more than 500,000 additional deaths in sub-Saharan Africa over the next year, bringing the region back to 2008 AIDS mortality levels. Even a 20% disruption could cause an additional 110,000 deaths.
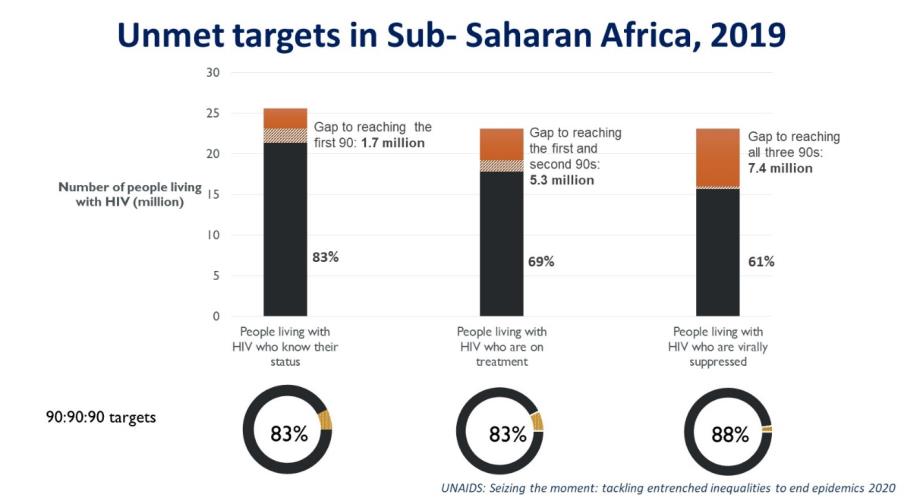
COVID-19 contributes to ART drug stockouts, reduces medical staff availability for HIV care, demotivates HIV patients to report at clinics, increase stigma and thus will result in additional HIV drug resistance (HIVDR). To make matters worse, testing for HIVDR is expensive, complex and requires sophisticated lab staff and equipment. However, countries require insights in the level of HIVDR in their populations in order for policy makers to make the right decisions for purchasing HIV drugs. Today, such information is obtained through expensive and laborious national HIV drug resistance monitoring and surveillance approaches along the guidelines of the World Health Organization (WHO). WHO monitoring and surveillance for HIVDR currently involves laborious and expensive DNA sequencing work on hundreds of individual patient samples that are carefully collected all over the country. Typically, such surveys cost in the order of 100,000 euro and take a year to perform.
In the current project, a radical simplification is proposed: instead of working on individual samples of patients, we mix these samples into one single ‘soup’. Subsequently we use modern DNA deep-sequencing techniques and powerful statistics to draw conclusions on HIVDR within the pertinent mix. With continuously decreasing prices of DNA sequencing and statistical analytics, the price for such an approach will be substantially lower (in the range of 10x) and results can be obtained in 1-2 months.
If proven of sufficient sensitivity and specificity, the ‘pooled HIVDR monitoring approach’ might become the new standard worldwide.
This project is an example of JLI taking risks and opposite approaches: ‘how can the good turn into the best’? Such an approach is typical for one of the aspects of the overall strategy of JLI, which is to seed-fund daring global health projects that may be controversial and less eligible for funding by established funding institutions. Such JLI seed-funding is meant to generate preliminary results, that can convince more conventional funders to take over and invest in the next phase(s) of the pertinent project. Criteria for such JLI funding include that the project should address a compelling global health problem, the solution should contain a substantial component of digital technology, the results of the project should in principle be more universally applicable.
This project is about a relatively simple and at first glance quite ‘sloppy’ biomedical assay and subsequent big data analytics that could make a true difference in the area of combating HIV. The project is performed in collaboration with renowned researchers from the Harvard School of Public Health, USA.
Partners